Introduction
- here we give a brief overview of GeneLynx purpose and organization.
Text search - if you want to locate a gene using a keyword or other identifier.
Quick Search - searches through all keywords and alphanumeric identifiers (including accession numbers). Simple, fast and powerful.
Advanced search - searches specified identifiers, logically combined in various ways.
NOTE TO THE USER - you can help us make it better
BLAST search - if you want to locate a gene in GeneLynx using a query nucleotide or protein sequence.
The contents of a GeneLynx record - on finding your way through the list of links for a particular gene.
User comment submission - what to do to help us improve the quality of GeneLynx data by correcting the errors you noticed and pointing to information that is missing.
Batch retrieval of GeneLynx IDs - now you can submit a list of identifiers (accession numbers, OMIM IDs, LocusLink numbers etc.) and GeneLynx will match them to the corresponding GeneLynx records. The resuld is a list in either HTML or plain text format which you can use for linking your database to GeneLynx.
Submitting a new resource for inclusion in GeneLynx - if you are in charge of a database with entries relating to human gene and would like GeneLynx to include links to it, we have tried hard to make it as easy as possible for both you and ourselves. Please read how to submit the data for your resource, and it will be included in GeneLynx in no time!
- Appendix 1: Resources linked to GeneLynx
- Appendix 2: Identifiers searchable by Quick Search
- Appendix 3: Identifiers searchable by Advanced Search
- Appendix 4: Browser compatibility issues
Introduction
GeneLynx is a service whose aim is to provide the fastest way between an IDENTIFIER for a given HUMAN GENE and all the data available for that gene on the web. It is organized as a set of HYPERLINKS leading to numerous databases and other resources. The number of currently linked resources is 32, and new ones are added regularly. For a complete list of linked resources, see Appendix 1.
An IDENTIFIER to search GeneLynx with is almost any piece of information that pertains to the gene of interest. It can be e.g.
a keyword, usually a word contained in the name or description of the gene, but also any alphanumeric identifier such as an accession number or ID (e.g. Swiss-Prot ID). The list of identifiers that can be used as keywords is given in Appendix 2. They can be used in either Quick Search or Advanced Text Search.
an identifier of the specified type, either numeric or alphanumeric, can be specified together with its type using the Advanced Text Search form. The list of identifiers that can be searched this way is given in Appendix 3.
a nucleotide or protein sequence can be used to locate a gene in GeneLynx system by searching them against cDNA and EST sequences associated with GeneLynx records. The searches are performed using BLAST 2.1.
Text search
There are two different ways of text search. In most cases the simpler Quick search will be more than enough powerful to locate a GeneLynx record for a gene of interest. Use Advanced search only in the cases where
it is not possible to formulate a query that is precise enough using Quick Search, or
you want to search GeneLynx by purely numeric identifiers, such as those of GDB, LocusLink, or OMIM, which are currently not searchable by Quick Search.
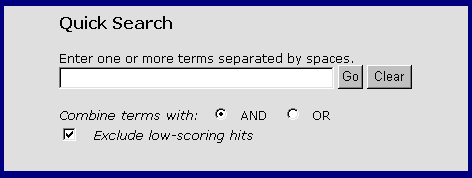
Quick Search
The Quick Search can be accessed from either GeneLynx Home Page or from Text Search page. There is no difference in function between the two.
"Combine terms with" option. Currently the query is separated into words, and some characters (punctuation, brackets, special characters) are ignored. Each word is then looked up in GeneLynx index table.
In the case the user has chosen to combine terms (words) by "AND", only those GeneLynx records containing all words from the query are displayed in the result list.
If the terms were combined by "OR", the list of hits contains GeneLynx records for genes that contain at least one of the terms in the query. Each of the terms is given the score value, which is inversely related to the number of GeneLynx records that share the term. For example the term "SYS_HUMAN" (a Swiss-Prot ID) is associated with only one GeneLynx record, and therefore has higher score than e.g. the term "clathrin" which in the current version of GeneLynx is associated with 21 records. The list of hits is sorted by decreasing sum of scores per record.
"Exclude low scoring hits" option. This option only affects searches performed with "Combine terms with OR" option. Some terms from the index are associated with a large number of GeneLynx entries, e.g. "protein" (9533) or "gene"(1895). Excluding low-scoring hits means excluding from the list the hits that contain only such frequent terms. For example, if the query was "divalent iron protein", at the top of the list there will be records that contain all three terms, followed by records that contain both "divalent" and "iron", then those that contain only "divalent", then those matching only "iron". If the low scoring hits were included, the list will not contain the records matching only "protein".
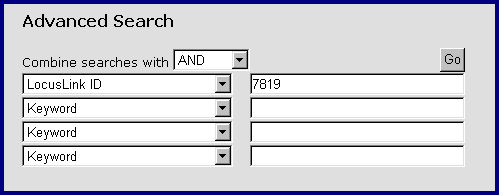
Advanced Search
The Advanced text search can be accessed from Text Search page. It is meant to be used in the cases when you can not formulate the query precisely enough by using Quick Search, or for searching specific numeric identifiers.
Advanced Search Form
The terms can be combined with "AND" (lists the records that contain all identifiers), "OR" (lists the records that contain either one of the given identifiers), or "BUTNOT" (lists the records that contain the first identifier, but none of the identifiers below).
NOTE TO THE USERS
The GeneLynx search systems described above are planned to evolve based on feedback provided by you. If you need a search feature that you know you or others would use often, please send your suggestions to Boris Lenhard. We feel that many of the more "intelligent" search algorithms are slow and unnecessarily fuzzy, retrieving too many irrelevant hits, so we would like our users to guide us in the right direction.
BLAST Search
GeneLynx provides a basic BLAST search utility specifically designed for retrieving GeneLynx records.
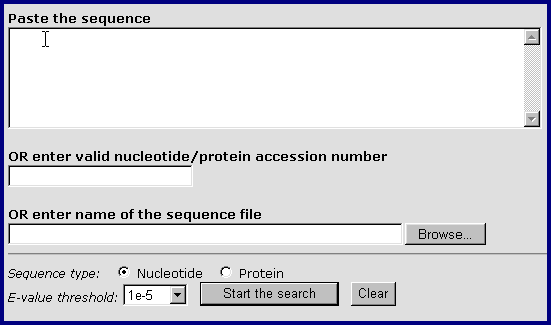
There are three ways to submit a sequence:
Paste the sequence: Paste the sequence from the clipboard. The sequence can be in raw (sequence only) or FASTA format (the ID line is simply ignored). It can also contain numbers and spaces (which are both ignored), which makes direct pasting of the sequence portions of GenBank/EMBL/Swiss-Prot records possible.
Enter valid nucleotide/protein accession number. The accession number of any sequence that can be retrieved from NCBI.
Enter the name of a sequence file. If the sequence that you want to submit is in a local file on your computer, enter its filename with the full path. The sequence can also be in raw or FASTA format. In case that the file contains multiple sequences in FASTA format, only the first one is used.
Other options available are:
Sequence type: Hopefully this is obvious. Soon the default type will be "Auto", i.e. GeneLynx will try to determine the type from the sequence itself.
E-value threshold: Basic option for regulating the threshold significance of BLAST hits. If there are users who think it could be controlled in a more adequate manner, please send your suggestions to Boris Lenhard.
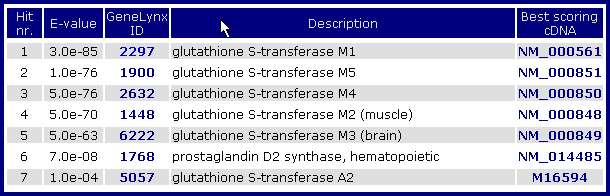
The submitted sequence is searched against a set of (1) all cDNAs associated with GeneLynx records, and (2) assembled EST sequences of EST-only GeneLynx records. A typical list of hits looks as follows:
The hits are sorted by increasing E-value, with only one (best scoring) cDNA per GeneLynx record shown. The link to the NCBI record for best scoring cDNA sequence is given in the rightmost column.
The contents of a GeneLynx record
GeneLynx contains hyperlinks associated with a GeneLynx record which, as a rule, corresponds to one human gene. If you happen to find an entry that appears to violate this rule, you are strongly encouraged to submit a comment for that record.
Resource categories
The links point to different resources, which are divided into following resource categories:
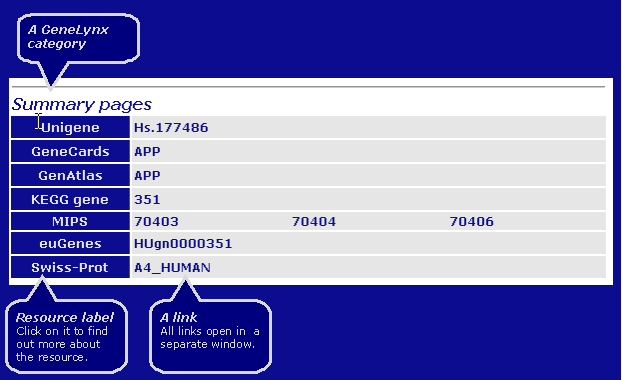
Summary pages - a somewhat arbitrary category, containing links to resources that provide a summary for the gene and/or extensive set of further links. Most of these are gene-based like GeneLynx itself. One notable exception is the Swiss-Prot, which is protein-based (also a member of "Protein sequences" category, making it the only resource tat is duplicated on the page), but which contains a wealth of links to other resources.
Genomic Resources - resources that provide information on the gene in the context of its location in the human genome. It includes resources with genomic sequences, chromosome maps etc.
Transcripts - collection of resources on mRNA/cDNA sequences.
Protein sequences - major protein sequence databases.
Protein structure and domains - a collection of links on protein tertiary structure, protein domains and patterns.
Protein function and disease links - this is a category that will probably be split in two in future releases of GeneLynx. It contains link to resources on enzyme function, metabolic pathways and disease associations of a given gene product.
Homologs - this is a growing category of links to information on nonhuman genes and proteins homologous to the gene of current GeneLynx record.
ESTs -although ESTs are also transcribed sequences, their abundance and chaotic content lead us to the decision to put them in a separate group. It contains links to EST sequences and assembled EST clusters.
The categories are subject to change (as is anything else, if necessary) .
Information available for the links
The layout of a GeneLynx category is:

Since GeneLynx is envisioned as a collection of links, and not a summary of textual information about the gene, current version does not contain descriptions of the links' content. Instead, links open in separate windows so that several resource pages can be inspected simultaneously while retaining access to other links.
Note: Once GeneLynx reaches Release 1.0 it will also contain "mouse-over" descriptions of link contents (i.e. upon placing mouse pointer on e.g. a Swiss-Prot link, the contents of the Swiss-Prot record's DE field will appear in browser status line, and above the link itself in browsers supporting that feature). We would appreciate user's feedback on potential usability of that feature, since it is not too trivial to implement.
User Comment Submission
We believe that user comment submission system is one of major features that makes GeneLynx more usable than similar resources. It is simple and self-explanatory, but here is a quick tour of its features.
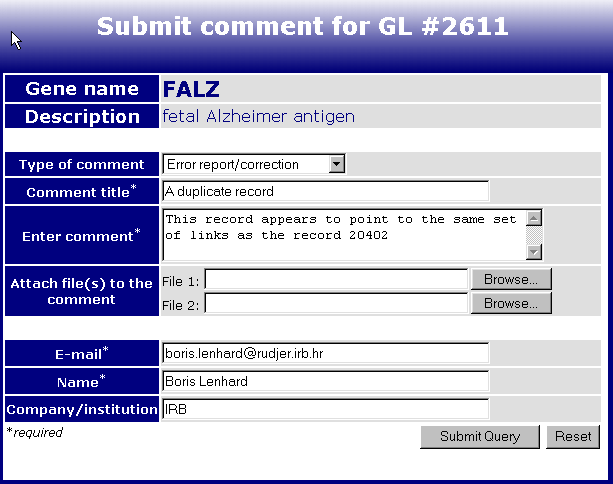
The comment submission form is accessed by clicking on the links that looks like this:
and is located immediately below the name and the locus of the gene. By clicking on the above link on the page of GeneLynx record #2611, you access its comment submission form:
The fields are:
Type of comment - Choose among 'Error report/correction', 'Supplementary information' or 'Other'. This field is for curator's orientation only.
Comment title - Here you state the reason for your comment.
Enter comment - Here you type the text of the actual comment. You are encouraged to be as detailed as possible here, for that will make curator's life easier. Feel free to include web addresses and literature references that support your comment, if the problem is not trivial.
Attach file(s) to the comment - along the lines of being as detailed as possible, you may want to include some files (documents, alignments, BLAST search reports etc.) that corroborate the claim you make in the comment.
E-mail , Name, Company/Institution - I do not need to explain those, do I :)
Remember, all of the form fields are there so that the curator could make corrections more quickly. In a great majority of cases the curator will be much less knowledgeable about a particular gene than the person who sets out to send a comment on it.
When you submit a comment, you will receive an E-mail message confirming it. The comment will simultaneously appear in the bottom if the Comment form page, so that other GeneLynx users will be able to read it (and all the previous comment on that record) before submitting their own. In the above example (#2611), there were no previous comments, so that after our comment submission and subsequent access to the #2611 comment page, the bottom of the page looked like this:
In the above example, two records with the same set of links have been detected, which usually means that GeneLynx clustering algorithm has made a rare error of not grouping all cDNAs of the FALZ gene into a single cluster. A GeneLynx curator will inspect the cDNAs for both GeneLynx records (#2611 and #20402), and if they really belong to the same gene, merge them into a single one. In this case, the merged record would be #2611, and #20402 would be retired. If the user accesses #20402 page later (e.g. via a bookmark made previously), she will access a page with retirement notice and a link pointing to the #2611 record.
Linking to GeneLynx - batch retrieval of GeneLynx IDs
(This service is currently experimental. We need your feedback.)
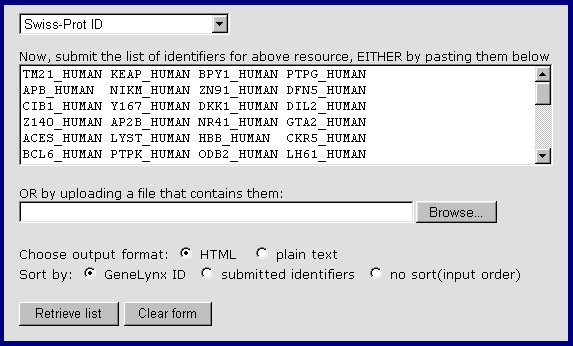
This service might be of interest to you in the following cases:
Suppose you have performed a search of a database or an experiment including large number of nucleotide or protein sequences, and obtained a list of identifiers (nucleotide accession numbers, protein sequence identifiers, HUGO numbers of genes etc.) GeneLynx enables you to paste or upload this list to the Batch GeneLynx page and obtain a list in which these identifiers are associated with appropriate GeneLynx IDs and descriptions.
You have a list of identifiers from a resource that is included in GeneLynx, and want to retrieve a list of GeneLynx numbers for linking from your resource's web pages.
In either case, all you need is a list of identifiers (nucleotide accession numbers, protein sequence identifiers etc.) that you submit by pasting or uploading it on the "Linking to GeneLynx" page. E.g. if you have a collection of Swiss-Prot IDs:
Paste the IDs (or select a plain text file containing the IDs to upload). Select output format, and the sorting criterion. (If you choose to sort by submitted identifiers, and the submitted identifiers are numbers, a numerical sort is automatically performed; otherwise the identifiers are sorted alphabetically).
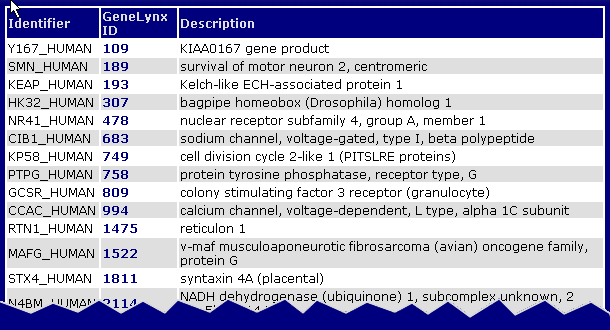
The HTML output looks like this:
It is meant for browsing, and provides descriptions and links to the correspondig GeneLynx records.The HTML page can also be saved locally for later use.
The text format is a two-column, tab-delimited format that is easily parsed by computer programs or loaded into a spreadsheed or database table:
- Y167_HUMAN 109
- SMN_HUMAN 189
- KEAP_HUMAN 193
- HK32_HUMAN 307
- NR41_HUMAN 478
- CIB1_HUMAN 683
- KP58_HUMAN 749
- PTPG_HUMAN 758
- GCSR_HUMAN 809
- CCAC_HUMAN 994
- RTN1_HUMAN 1475
- MAFG_HUMAN 1522
- STX4_HUMAN 1811
- N4BM_HUMAN 2114
...
Submitting a new resource for inclusion in GeneLynx
(This service is currently experimental. We need your feedback.)
To facilitate (and encourage) addition of new database resources to the collection of GeneLynx links by their authors and curators, we have developed a (highly experimental) standardized procedure for external resource submission. Currently, a resource to be submitted must meet the following requirements:
Its individual records must be accessible on the web via a URL that includes their unique identifier.
each unique identifier should be associated with an identifier belonging to a resource that is already included GeneLynx. The associations should be listed in a two-column plain text file (tab-, space- or comma-delimited).
- NEW001 2049
- NEW002 79154
- NEW003 7067
- NEW004 64979
- NEW005 55438
- NEW006 5324
- NEW007 57528
- NEW008 55116
- NEW009 215
- NEW010 10421
- NEW011 29056
- NEW012 10072
- NEW013 56617
- NEW014 29108
- NEW015 7442
...
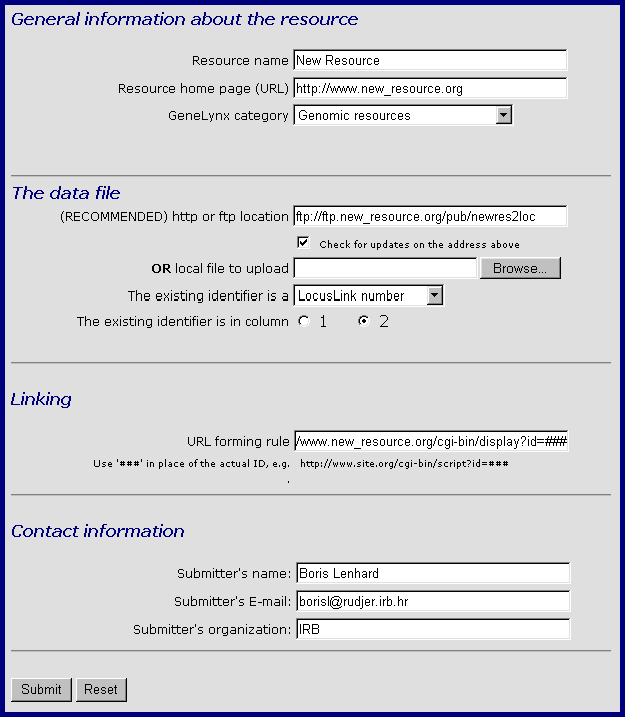
In the above case, the second column contains LocusLink IDs (the order of columns can be reversed). Now go to GeneLynx resource submission page and fill the submission form as follows:
General Information about the resource is necessary for the proper labelling and categorization of the new resource.
Resource name is the name that will appear in the resource label on a GeneLynx record page.
Resource home page is the URL of the new resource's home page, preferably with additional information about the resource.
GeneLynx Category is the label for a group of related resources (see The structure of a GeneLynx record).
The data file section is where you specify the location of the newres2loc file. You can either specify its URL on a http or anonymous ftp server, or upload it from your local file system. However, we recommend that you specify the URL, preferably with the "Check for updates" option. That the file will be reloaded for each GeneLynx update, so you can just keep the file current instead of resubmitting it to geneLynx each time it is changed.
In "The existing identifier is a" menu, choose the identifier to which your resource's identifier is matched in the file you are submitting (in the above case "LocusLink number").
The "Existing identifier is in column" should be obvious. In our example, the existing identifier is LocusLink number which is in column 2 of the newres2loc file.
Linking section is where you type the URL forming rule for retrieving records of the new resource. Type triple hash ### where the actual identifier should be inserted.
In Contact information, your E-mail is required in order to recieve the confirmation of successful submission and final inclusion in GeneLynx by the curator. It is also possible that a GeneLynx curator coctacts you for additional information.
After you submit the required information, you will come to Submitted resource test page which looks like this:
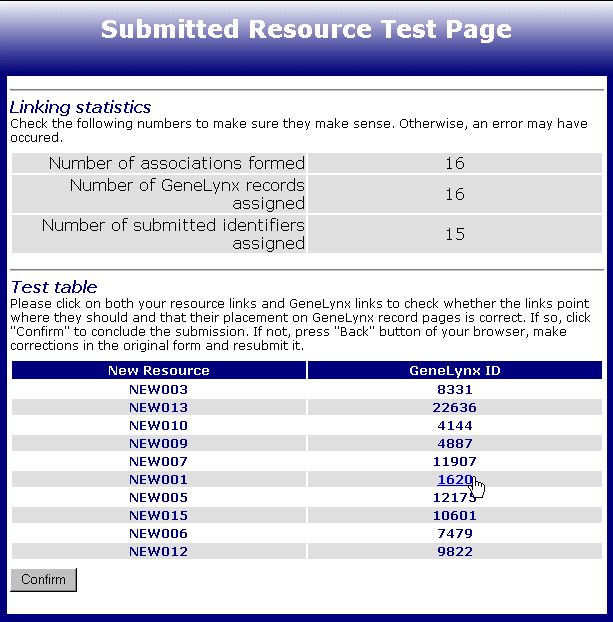
Under the table containing some simple figures about the submission (you should check them to make sure they make sense), there is the Test table with up to 10 randomly picked pairs of newly formed associations between new resource identifiers and GeneLynx records. There are three things you could check:
that the new resource identifiers link to the proper web addresses; click on them to open the corresponding web pages in a new window
that the selected new resource identifiers are associated with appropriate GeneLynx records;lick on a GeneLynx ID in the second column to open the GeneLynx record
after you open the GeneLynx record page, locate the new resource on the page. It should not be too hard - its Resource label is the only one coloured red:
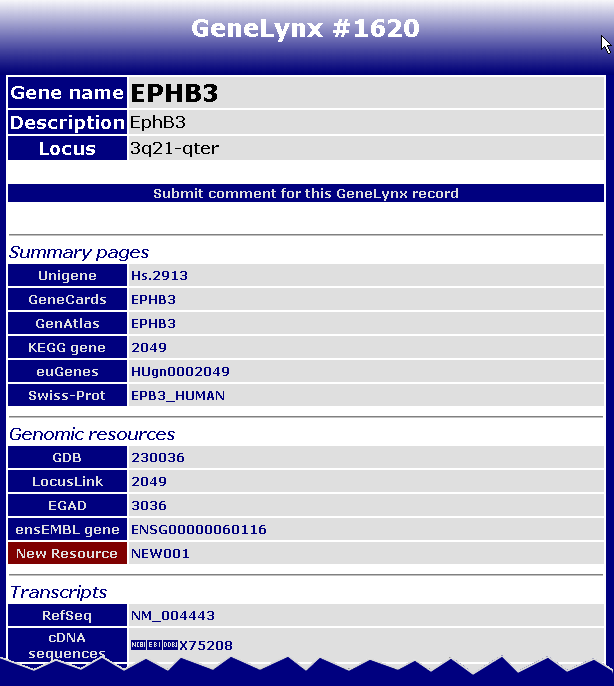
Check that it is in the desired category and that the links work as they should.
If everything is in order, click "Confirm"on the Submitted resource test page, and you are done. You will receive an E-mail message confirming the succesful submission, and a GeneLynx curator will notify you of further developments.
Appendix 1
- Resources linked to GeneLynx
- Gentaur Group
- UniGene Home page
- LocusLink
- GeneCards Home page · About
- Swiss-Prot
- KEGG gene Home page
- EGAD Home page
- euGenes
- MIPS Home page · About
- HumanPSD Home page
- Genomic resources
- Genomic sequences
- GDB Home page
- GenAtlas Home page · About
- Ensembl gene Home page
- Transcripts
- RefSeq About
- cDNA sequences
- Ensembl transcript Home page
- Protein sequences
- Swiss-Prot Home page
- TrEMBL Home page
- PIR Home page · About
- GenPept
- Protein structure and domains
- PDB
- Closest PDB structure A link to the most similar available PDB structure for a given Genelynx gene product, as determined by HSSP.
- HSSP
- InterPro
- PRINTS
- PFAM
- BLOCKS
- SBASE
- PROSITE
- Protein function and disease links
- GeneOntology (at MGI)
- MEROPS proteases
- ENZYME database
- WIT
- BRENDA
- OMIM
- GeneClinics
- Networks and Pathways
- KEGG pathway
- PubGene
- Homologs
- Nucleotide
- Protein
- Unigene
- LocusLink
- MGD
- ESTs
- STACK cluster
- EST sequences
Appendix 2
Identifiers searchable by Quick Search
Identifier Remark
Keyword Any word from description/definition lines of the UniGene, LocusLink, SwissProt, Trembl and HUGO records associated with GeneLynx records
Nucleotide accession number GenBank, EMBL, DDBJ and EST sequences. Human only. If the query returns no hits, try a BLAST search with that accesion number.
Protein accession number Swiss-Prot, TrEMBL, PIR and GenPept sequences.If the query returns no hits, try a BLAST search with that accesion number.
Swiss-Prot ID e.g. SYS_HUMAN
PIR ID not the sama as PIR accession number
PDB ID
UniGene ID for Quick search, use the full identifier, e.g. Hs.4888
HUGO symbol Official human gene symbols
Other gene symbols Gene symbols other than the official (HUGO) ones, but used in literature and databases.
Appendix 3
Identifiers searchable by Advanced Search
Under construction...coming soon.
Appendix 4
Browser compatibility issues
GeneLynx has been tested on several platforms with different web browsers. While we tried to make the interface as browser-independent as possible, some browser just do not allow that. However, all the detected incompatibilities are of the cosmetic nature, making the pages less pretty than we intended it to be :) without affecting access to any of the GeneLynx capabilities. Here is a table that summarizes our experience:
Browser Version OS Known problems
MS Internet Explorer 4.0, 5.0, 5.5
MS Windows
(9x, NT,2000)
None
Netscape 6.0
Linux 2.2.17
Linux 2.4.4
None
Netscape Communicator 4.5, 4.61, 4.74
Linux 2.2.x
Tru64 Unix (Compaq)
The page layout is altered incorrectly upon window resizing
Mouse-over underlining of links does not work
There are no boxes around items in the left-side menu on all pages
Konqueror
2.1.1
2.1beta2
Linux 2.2.x
Tru64 Unix (Compaq)
Occasional minor occurrences of incorrect font attribute rendering; otherwise much better and faster than Netscape 4.X
Lynx Several
Linux 2.2.x
Tru64 Unix (Compaq)
Just for the those of Spartan spirit. Most features work, but the layout is hard to follow (No. we do not intend to work toward improving it - we strongly believe that a really negligible number of Lynx users will have ever heard of GeneLynx and vice versa.)
If you experience problems other than those listed in the table, please let us know.
Send comments and questions to Boris Lenhard